Authors
Gregory J. Berry
Columbia University Vagelos College of Physicians and Surgeons
New York-Presbyterian—Columbia University Irving Medical Center
New York, NY, United States
Tulip A. Jhaveri
Department of Internal Medicine
Division of Infectious Diseases
University of Mississippi Medical Center
Jackson, MS, United States
Paige M.K. Larkin
University of Chicago Pritzker School of Medicine
NorthShore University Health System
Chicago, IL, United States
Heba Mostafa
Johns Hopkins School of Medicine
Department of Pathology
Baltimore, MD, United States
N. Esther Babady
Clinical Microbiology and Infectious Disease Services
Department of Pathology and Laboratory Medicine and Department of Medicine
Memorial Sloan Kettering Cancer Center
New York, NY, United States
Endorsed By
.png?h=131&w=200&hash=D57AAF707D9E27DFA8E80AA3D8123086)
Respiratory viral infections are among the most frequent infections experienced worldwide. The COVID-19 pandemic has highlighted the need for testing and currently several tests are available for the detection of a wide range of viruses. These tests vary widely in terms of the number of viral pathogens included, viral markers targeted, regulatory status, and turnaround time to results, as well as their analytical and clinical performance. Given these many variables, selection and interpretation of testing requires thoughtful consideration. The current guidance document is the authors’ expert opinion based on the preponderance of available evidence to address key questions related to best practices for laboratory diagnosis of respiratory viral infections including who to test, when to test, and what tests to use. An algorithm is proposed to help laboratories decide on the most appropriate tests to use for the diagnosis of respiratory viral infections.
INTRODUCTION
Community-acquired respiratory tract infections occur with varied but predictable frequency throughout the year (1). Respiratory tract infections caused by viral pathogens are among the most common reasons for healthcare visits (2). Transmission occurs through multiple routes including directly through contact or inhalation of viral particles, or indirectly through routes such as contact with contaminated fomites and self-inoculation of the respiratory tract mucosa (3, 4).
The most common viruses causing respiratory illness include picornaviruses (rhinoviruses and enteroviruses), influenza viruses (influenza A and influenza B), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), respiratory syncytial virus (RSV), parainfluenza viruses (PIVs), endemic human coronaviruses, adenoviruses, and human metapneumovirus (hMPV) (5). Respiratory viruses such as bocavirus and Middle East respiratory syndrome coronavirus (MERS-CoV) are less common. Characteristics of these viruses, including associated clinical syndromes, strains/types, and seasonality can be found in Table 1.
The signs and symptoms of respiratory illness overlap for most respiratory viruses and may be indistinguishable from bacterial infection on clinical presentation. Clinical syndromes associated with respiratory viruses span a wide range of presentations from the “common cold” to bronchitis and severe pneumonia (Table 1). Acute upper respiratory tract infections are generally mild and self-limited; however, severe complications may develop in both healthy individuals and in those with underlying conditions, which will be addressed in detail later in this section. Most severe complications relate to progression to the lower respiratory tract and include pneumonia, respiratory failure, and superimposed bacterial infection (6, 7).
Noninfectious complications including cardiac (e.g., acute myocardial infarction, myocarditis) and neurological (e.g., seizure, encephalopathy) complications resulting in increased mortality have been associated with influenza and other respiratory virus infections (8, 9).
The wide range of presentations may be due to differences in the host, the site of the infection, and the presence of additional pathogens. Both the innate and adaptive components of the immune systems are involved in the host defenses against viral infections (10, 11). Prior exposure to viral pathogens might provide partial immunity only, given the large diversity of circulating viral serotypes and genotypes and the frequent occurrence of mutations in viral genomes (12, 13).
Additionally, certain populations are more susceptible to severe disease, including children, older adults, and patients with underlying diseases, or suppressed immune functions (14). In children, particularly infants, the increased risk is due in part to their immature immune system which is characterized by a lack of immune memory and reduced innate and adaptive immunity (15). In children undergoing treatment for hematologic malignancies, infections due to influenza, parainfluenza, and RSV are particularly common and can result in increased morbidity (16, 17). In patients with underlying conditions, a defective innate immune system, including decrease (e.g., mucositis) or increase in mucociliary escalator function (mucus hypersecretion), may result in decreased clearance of viral pathogens and increased risk of infections (10). In adults >65 years of age, waning innate and acquired immunity associated with decreased function of memory cluster of differentiation (CD)-8+ T cells results in increased susceptibility to viral infections and severe disease (18, 19). Early in the COVID-19 pandemic, older age was identified as a significant risk for increased mortality and hospitalization due to SARS-CoV-2 (20).
Rapid microbiological diagnosis of acute respiratory infections has both therapeutic and prognostic implications. Knowledge of the virus epidemiology and a complete history and physical examination may assist in determining the most likely responsible pathogen (21). While healthy, immunocompetent hosts usually recover from acute respiratory infections without the need for laboratory diagnosis or treatment, specific diagnosis in immunosuppressed patients and those with underlying conditions is necessary for implementing appropriate, targeted therapy when available. Laboratory testing is necessary because the accuracy of clinical diagnosis alone is limited. In one study, clinical diagnosis of pneumonia based only on clinical findings such as fever, cough, sputum production, abnormal chest auscultation, and dyspnea had limited value in differentiating infectious vs noninfectious pneumonia, and bacterial vs viral pneumonia (22). Imaging studies, specifically chest X-rays and computer-tomography (CT) scans are necessary to diagnose pneumonia. However, imaging studies are not specific enough to definitively distinguish between viral and bacterial infections (23). The addition of procalcitonin and C-reactive protein significantly increased the sensitivity of clinical diagnosis from an area under the curve (AUC) of 0.79 (95% CI, 0.75–0.83) when all clinical findings were combined to an AUC of 0.92 (95% CI, 0.89–0.94); P < 0.001 (24). While these biomarkers improve the accuracy of clinical assessment for pneumonia, the specific etiology remains unknown. This limitation of clinical diagnosis alone or in combination with imaging and other biomarkers underscores the need and the importance of viral diagnostics for the rapid detection of respiratory viruses.
| Table 1. Common viral pathogens and associated clinical syndromes | |||||
| Family | Virus | Genomea | Respiratory clinical syndromes | Seasonality | Serotype/Strain |
| Adenoviridae | Adenoviruses (AdV) | dsDNA | Pharyngitis, common cold, laryngitis, bronchitis, bronchiolitis, pneumonia |
Circulate year-round |
7 species (A-G): >100 types. |
| Parvoviridae |
Bocaviruses (BoV) |
ssDNA |
Common cold, laryngotracheobrochitis (croup), bronchiolitis |
Winter/early spring |
HBoV-1: serotype associated with respiratory symptoms |
| Coronaviridae |
Endemic seasonal coronaviruses (CoV) |
ssRNA |
Common cold, pharyngitis, laryngitis, bronchitis, bronchiolitis |
Winter in temperate seasons |
NL63, OC43, HKU1, 229E |
| Severe acute respiratory syndrome coronaviruses (SARS-CoVand SARS-CoV-2) | ssRNA |
Severe acute respiratory syndrome; COVID-19: ranges from asymptomatic, mild to fatal |
Year-round for SARS-CoV-2 at this time |
SARS-CoV; SARS-CoV-2 |
|
| Middle East respiratory syndrome coronavirus (MERS-CoV) |
ssRNA |
Middle East respiratory syndrome (MERS), ranges from asymptomatic to mild to fatal |
Varies. Some studies report winter and summer months peaks. Others show no seasonality |
||
| Picornaviridae |
Enteroviruses (EV) |
ssRNA |
Common cold, pharyngitis, laryngotracheobrochitis (croup), hand-foot-and-mouth disease, bronchiolitis and pneumonia |
Peak incidence in summer/fall in temperate regions and rainy season in tropical |
EV-A, EV-B, EV-C EV-D: Notably, EV-D68 which is associated with flaccid myelitis |
| Human rhinoviruses (RhV) |
ssRNA |
Common cold, pharyngitis. Severe manifestations: otitis media, bronchiolitis, croup, and exacerbation of asthma with significant wheezing in children |
Year-round with peak incidence of September |
>100 serotypes |
|
| Paraxmyxoviridae |
Human parainfluenza viruses (HPIV) |
ssRNA |
Common cold, pharyngitis, croup. Severe manifestations: bronchiolitis, tracheobronchitis, pneumonia, and febrile and afebrile wheezing |
Biennialfall epidemics (HPIV-1/2); spring and summer epidemics (HPIV3) |
HPIV-1, HPIV-2, HPIV-3, and HPIV-4 |
| Orthomyxoviridae |
Influenza viruses (Flu) |
ssRNA |
Influenza, bronchitis, bronchiolitis, pneumonia, common cold, pharyngitis, laryngitis, croup |
Winter in temperate regions |
FluA (H1N1, H3N2, 2009 H1N1), FluB, FluC |
| Pneumoviridae |
Respiratory syncytial virus (RSV) |
ssRNA |
Bronchiolitis, pneumonia, common cold, croup, bronchitis, otitis media |
Seasonal epidemics in the winter (temperate regions) and in the late summer (tropical regions) |
RSV-A and RSV-B |
| Human metapneumovirus (hMPV) |
ssRNA |
Bronchiolitis, common cold, laryngitis, bronchitis, pneumonia |
Late winter and spring in temperate regions but with less predictability in tropical regions |
||
| Abbreviations: ss, single-stranded; ds, double-stranded. | |||||
WHAT TESTS SHOULD BE USED TO DETECT RESPIRATORY VIRUSES?
Several testing methodologies are available for the detection of respiratory viruses. They differ on several counts including the viral target (e.g., protein, DNA, RNA), performance characteristics, sample type used, level of complexity, and regulatory classification (Table 2). Respiratory viral testing can be performed in centralized laboratories, at the point of care, or outside of a healthcare setting. The different settings dictate the type of tests that can be utilized and are based on the complexity of the selected test as determined by the FDA during the premarket approval process. Examples of commercially available in vitro diagnostic (IVD) tests are listed in Table 3.
Sample Types for Respiratory Virus Testing
Specimen types are dictated by the acceptability requirements of FDA-approved assays. Some assays list NPSs, nasal swabs, throat swabs, or bronchoalveolar lavage fluids as acceptable specimens depending on whether the assay is used for upper respiratory tract infections (URTIs) or lower respiratory tract infections (LRTIs). The patient age may also play a role in sample selection, with nasal aspirates traditionally being the preferred samples type for young pediatric patients (28). Studies have shown that regardless of source, children have a higher viral load than adults and subsequently viral detection in this patient population tends to be higher (25). Patients over the age of 65 years may also exhibit decreased secretions and nasal dryness, further reducing the specimen quality (26). Patient comfort and compliance should be considered as well—if a patient is resistant to sample collection by NPS due to discomfort, then the sample quality and subsequent sensitivity could decrease as a result of suboptimal collection. The type of swab also impacts accuracy with one study finding that flocked NPSs in universal transport media have superior accuracy compared to NPSs using Dacron swabs (27).
Complexity and method of test will predetermine acceptable specimens. When considering specimen sources, a trade-off between timeliness, quality of specimen, and sensitivity of the method used on the specimen should be evaluated (25). Non-NPS upper respiratory sources (e.g., lateral mid-turbinate nasal, anterior nares, and saliva) can be collected by a wider range of healthcare personnel or even self-collected.
Methodologies Utilized for Detection of Respiratory Viruses
Viral antigen detection. One rapid option for detection of respiratory viruses is the use of antigen tests, which uses immunochromatographic methodologies such as lateral flow immunoassay (LFA) (29, 30). Currently, FDA-approved or FDA Emergency Use Authorization (EUA) antigen tests are only available for influenza A, influenza B, SARS-CoV-2, and RSV, limiting their applicability to the broad etiologies of respiratory infections (Table 2) (26, 29–33).
| Table 2. Methods used for routine detection of respiratory virusesa | |||||
| Viral target | Method | Specimen types | Pros | Cons | Regulatory status |
| Antigen | LFA |
- NS - NPS |
- Rapid - Inexpensive - Readily available - Instrument free |
- Lower sensitivity - Limited targets - Low throughput |
- CLIA-waived - POCT - Home test |
| DFA |
- NS - NPS - OPS - Sputum - BAL |
- Rapid - Can be multiplexed - Provides specimen quality check |
- Moderate sensitivity - Limited targets - Low throughput - Multiple steps - Manual - Subjective interpretation - Requires highly trained staff |
- Moderate complexity - High complexity |
|
| Nucleic acids | NAAT (e.g. PCR, MAP, TMA) |
- NS - NPS - OPS - Saliva - Sputum (pneumonia panels) - BAL (pneumonia panels) |
- Rapid (sample-to-answer platforms) - Highly sensitive - Can be highly multiplexed - Low-to-high-throughput options |
- Expensive - For high-and moderate-complexity tests, requires highly trained staff - Multiple steps for some platforms |
- CLIA-waived - POCT - Home test - Moderate complexity - High complexity |
| aAbbreviations: LFA, lateral flow immunoassay; DFA, direct fluorescence antibody; NAAT, nucleic acids amplification test; NS, nasal swabs; NPA, masopharyngeal swabs; OPS, oropharyngeal swabs; BAL, bronchoalveolar lavage; CLIA, Clinical Laboratory Improvement Amendments; POCT, point-of-care test. | |||||
Antigen point-of-care tests (POCTs) offer the advantage of being inexpensive, easy to perform with rapidly available results, and waived under the Clinical Laboratory Improvement Amendments (CLIA) regulations. Antigen POCTs can therefore be performed by nonlaboratory personnel and some are available for at-home testing (e.g., SARS-CoV-2 and influenza) (30). This is especially advantageous for areas where a centralized lab is not geographically close or a fast result would change management (e.g., an influenza-positive patient receiving Tamiflu) as most antigen tests take 10 to 15 min on average (30, 33). Moreover, one study demonstrated that antigen POCTs in the outpatient setting was able to predict influxes of emergency department (ED) cases of influenza, highlighting the versatile use of antigen testing (34). Unless a digital sensor is used for interpretation of antigen tests, no equipment is needed (30), allowing for use in small spaces and resource-limited settings.
Antigen testing does not amplify the target signal and thus requires a higher viral load for detection compared to nucleic acid amplification tests (NAATs) (30). Multiple studies have demonstrated that sensitivity is further decreased for adult populations compared to children, likely due to difference in viral loads in clinical samples (26, 29, 30, 35–37), especially with RSV. The lower sensitivity has been extensively studied during the SARS-CoV-2 pandemic. While antigen testing for SARS-CoV-2 was a useful screen, antigen testing was most accurate at high viral loads with lower positive samples often missed (38–40). The manual interpretation of antigen tests may be subjective and lead to inaccurate results. Some assays have reduced this issue by incorporation of a sensor that reads and interprets the results, rather than relying on visual inspection by a person, which may increase the test accuracy (29). Antigen testing is also unable to differentiate between subtypes of viruses, including influenza A (41, 42). However, the ability to differentiate subtypes of influenza A is not currently clinically relevant.
Viral antigens can also be detected using direct fluorescent antibody (DFA) assays in which the clinical sample is fixed to a solid surface and the viral antigen detected using a fluorescently labelled, monoclonal antiviral antibody. DFA assays are more labor-intensive than rapid LFA and are performed in high-complexity laboratories. DFA assays are available for select viruses, including RSV, influenza, and parainfluenza (26, 32, 37). Like LFAs, DFA assays are less sensitive compared to NAATs. During the 2009 H1N1 pandemic, influenza DFA assay was only 65% as sensitive as PCR, with some estimated ranges from 47.2% to 93% depending on patient population (43). Culture of DFA-negative specimens was able to detect 51% of the influenza cases that were missed by DFA assay alone (43).
Nucleic acids detection. NAATs are the gold standard for diagnosis of respiratory viruses (35) and can be modified to account for new strains and viral mutations (42). Several NAAT methodologies exist for nucleic acids detection of viruses including loop-mediated isothermal nucleic acid amplification (LAMP), transcription-mediated amplification (TMA), and real-time PCR (29, 41, 42). In the case of RNA viruses, a reverse transcription step is required to convert the RNA to DNA prior to amplification. NAATs do not offer information regarding infectiousness or active disease (32); patients may remain positive for days to weeks after disease resolution, discouraging the use of repeat testing of positive patients (44). With the exception of those approved for POCT use, most molecular assays are performed in high- or moderate-complexity laboratories and are typically associated with longer turnaround time (TAT) (31). An advantage of these assays is that they usually have a higher throughput and can be automated, which is particularly useful in high-volume settings (42). Automation further reduces contamination risk and human and pipetting error as well as reducing hands-on time required by staff, making this option appealing to many laboratories (42). There are a wide breadth of platforms, manufacturers, and panels available for molecular tests, with instrument overlap for non-respiratory testing. Some multiplex NAATs include both bacterial and viral targets and can include over 20 targets in one assay (Table 3) (31, 32, 35). This is particularly useful when the differential is broad or when viruses exhibit different irregular seasonality (31). While FDA-approved respiratory assays are widely available, some laboratories continue to utilize laboratory-developed tests (LDTs) due to cost, modifying FDA-approved assays (for example, bronchoalveolar lavage [BAL] validation), and customizability for tailoring testing for specific patient populations. Rare causes of respiratory viral infections are not commonly included on the commercial respiratory panels. For example, since 2012, MERS-CoV has caused over 850 deaths, a fatality rate of about 35% (45, 46). However, in the United States, only 2 patients tested positive for MERS-CoV; both cases occurred in 2014 in healthcare workers who traveled to Saudi Arabia (47). Given the low risk of MERS-CoV outside of the Middle East, many of the current diagnostic panels for routine, clinical use in the United States do not include a target for this virus.
| Table 3. Examples of molecular respiratory viral testing divided by the level of complexity (as of September 28, 2022) | |||
| Waived | Nonwaived | ||
| At home | Point of care | Moderate complexity | High complexity |
| Cue COVID-19 Test for Home and Over the Counter (OTC) Use | Abbott Diagnostics/ID NOW COVID-19 |
ARIES® Flu A/B & RSV Assay |
Abbott Molecular/Alinity m SARS-CoV-2 |
| Detect Covid-19 Test |
Accula Flu A/Flu B Test |
BioFire COVID-19 Test 2 |
Abbott Molecular/RealTime SARS-CoV-2 assay |
| Lucira CHECK-IT COVID-19 Test Kit |
Accula RSV Test |
BioFire Respiratory Panel2.1 (RP2.1) |
artus Infl A/B RG RT-PCR Kit |
| Accula SARS-CoV-2 Test |
ePlex Respiratory Pathogen Panel |
BioCode Respiratory Pathogen Panel (RPP) |
|
| Alere i NAT Flu A/B |
Focus Simplexa Flu A/B & RSV Direct |
BioFire FilmArray Pneumonia Panelplus |
|
| cobas Influenza A/B & RSV Nucleic Acid Test for Use on the cobas Liat System |
Nanosphere Verigene System (Multiplexed Nucleic Acid Test Detecting 13 Respiratory Viruses and 3 Bordetella Species) |
CDC Human Influenza Virus Real-time RT-PCR Diagnostic Panel, Influenza A/ B Typing Kit, CDC Human Influenza Virus Real-time RT-PCR Diagnostic Panel, Influenza A Subtyping Kit, CDC Human Influenza Virus Real-time RT-PCR, Influenza A/H5 Subtyping Kit |
|
| Visby MedicalCOVID-19 Point of Care Test |
eSensor Respiratory ViralPanel(RVP) |
||
| Visby MedicalCOVID-19, influenza A and influenza B Point of Care Test |
|||
| Xpert Xpress CoV-2 plus |
QIAstat-Dx Respiratory Panel |
Focus Diagnostics Simplexa FluA/B & RSV |
|
| Xpert Xpress CoV-2/Flu/ RSV plus |
GEN-PROBE Prodesse ProFAST + Assay (SeasonalInfluenza A/H1 A/H3 2009 H1N1 Influenza Viruses) (Seasonal Influenza A/H1 A/H3 2009 H1N1 Influenza Viruses Multiplex real-time PCR) |
||
| Xpert Xpress Flu/RSV |
Hologic/Aptima® SARS-CoV-2 Assay |
||
| Hologic/Panther Fusion SARS-CoV test |
|||
| IMDx Flu A/B and RSV for Abbott m2000 | |||
| JBAIDS Influenza A Subtyping Kit |
|||
| JBAIDS Influenza A&B Detection Kit |
|||
| Luminex NxTAG Respiratory Pathogen Panel on MAGPIX |
|||
| NeuMoDx™ SARS-CoV-2 Assay |
|||
| QUIDEL MOLECULAR INFLUENZA A + B |
|||
| ASSAY |
|||
| QUIDEL MOLECULAR RSV + HMPV |
|||
| ASSAY |
|||
| Roche/Cobas®SARS-CoV-2 Test |
|||
| aSeveral SARS-CoV-2 tests which were initially FDA-EUA are now fully FDA-cleared (e.g., the BioFire RP 2.0, Panther Fusion SARS-CoV-2/Flu A/B/RSV Assay and the Roche cobas SARS-CoV-2). Most up-to-date information can be found at https://www.accessdata.fda.gov/scripts/cdrh/devicesatfda/index.cfm |
|||
In 2015, the first CLIA-waived POCT NAAT for influenza was approved (30). There are now multiple EUA or FDA-approved POCT NAATs available for influenza A, influenza B, RSV, and SARS-CoV-2 with results available within an hour (29, 31). Not all POCTs are CLIA-waived and examples of waived and non-waived assays are shown in Table 3. In addition to rapid TAT, these assays offer the advantage of often being closed systems with minimal hands-on time and sample-to-answer format. However, given the increased sensitivity of molecular methods and the utilization of such assays in nonlaboratory settings by personnel not trained in molecular techniques, environmental contamination is a risk and mitigation measures should be considered to prevent false-positive results (30).
Next-generation sequencing (NGS) is another nucleic acid-based methodology that can be used to identify and characterize respiratory viruses. As there are currently no FDA-approved NGS assays for respiratory infections, availability and widespread use is limited and currently offered NGS assays are LDTs primarily used for viral genome characterization and not diagnosis (48).
Replicating virus detection. Viral culture has mostly been replaced by antigen and molecular testing in most clinical laboratories. However, viral culture offers the advantage of providing information regarding recovering infectious virus and phenotypic drug susceptibility, but it requires specialized facilities and poses biosafety concerns (26). There is no single universal cell line for testing of respiratory viruses, meaning that there must be a degree of clinical suspicion when ordering and particular cell lines must be used depending on the suspected virus (26). Moreover, many respiratory viruses are difficult to isolate or are slow-growing and the TAT for viral culture may be too long (up to 14 days) to be clinically useful (26, 35). Nevertheless, viral culture is particularly useful for phenotypic drug resistance and vaccine development in the case of influenza (42). Viral culture is less sensitive than PCR for the detection of viruses in clinical samples (42). In the case of SARS-CoV-2, one study found that only 46.4% of Reverse- Transcriptase (RT)-PCR or antigen-positive SARS-CoV-2 specimens had recoverable virus, with a higher success rate with lower threshold cycle (Ct) values (38).
Viral antibodies. Serology is rarely clinically used for diagnosis of respiratory infections and is not widely offered for most respiratory viruses. The assessment of IgG can be confounded by prior infection or vaccination depending on the virus (26). Acute and convalescent samples are often required to help establish active or recent infection, which requires additional collection and delayed TAT, making the test rarely clinically actionable (32). SARS-CoV-2 has highlighted the utility of serology and immunological assays for assessment of population seroprevalence, potentially establishing previous infection, and determining immune recognition, whether due to immunization or previous exposure (49, 50). Taken together, serology remains a key tool in epidemiological investigations but rarely plays a role in clinical diagnosis of acute viral respiratory infection.
Recommendations
- Testing should be performed following FDA-approved manufacturers’ recommendations.
- The preferred specimen type for URTI is an NPS. Alternatives when an NPS is not practical includes nasal washes (pediatrics), mid-turbinate swabs, nasal swabs, throat, or saliva as validated.
- For infections in the lower respiratory tract, BAL can be used.
- The preferred method for diagnosis is an NAAT. When an NAAT is not readily available, antigen tests could provide an alternative; however, they have lower sensitivity compared to NAATs.
- DFA, serology, and viral culture are not recommended for routine diagnosis.
HOW SHOULD THE RESULTS OF RESPIRATORY TESTING BE INTERPRETED?
Interpretation of positive or negative test results depends on the pretest probability of viral infection, which can be influenced partly by epidemiological factors, which can be determined by accessing current national (e.g., CDC website) and state epidemiologic data (e.g., state-specific public health website), and clinical factors including typical signs and symptoms of infections. The positive predictive value (the probability of disease given a positive result) and the negative predictive value (the probability of absence of disease given a negative result) take into consideration the prevalence of the circulating virus and the specificity of the various tests should be considered for accurate interpretation of tests results. For patients with low pretest probability of infection, e.g., an asymptomatic person in a low-prevalence setting, there is a higher chance of a false-positive test. In this situation, a single negative rapid diagnostic test is sufficient to rule out infection, while a positive test may require a repeat test for confirmation. For patients with high pretest probability of infection, e.g., a symptomatic person in a high-prevalence setting, there is a lower chance of false-negative test. Hence, a single positive rapid diagnostic test is sufficient to rule in infection prompting isolation, notification of providers and contacts, and consideration of treatment, while a negative test would prompt a repeat test in 2 to 3 days to exclude viral infections (51).
Interpreting Molecular Test Results
A positive molecular result for a respiratory virus usually confirms an infection with the virus but does not mean the presence of infectious virus. It is important to note that the molecular detection of respiratory viruses is a sensitive approach that is capable of detecting nucleic acids prior to symptoms and for extended periods of time after symptoms clear. Presymptomatic and asymptomatic positive SARS-CoV-2, influenza, and other respiratory viruses are common and correlated with recovery of infectious virus and possible viral transmission (52–55). Prolonged shedding of viral nucleic acid has been reported in asymptomatic patients recovered after infection with SARS-CoV-2 (56–59), influenza (60, 61), and other respiratory viruses (62). Additionally, prolonged respiratory viral shedding for weeks to months was observed in immunocompromised patients (63–66). Prolonged viral detection could challenge the interpretation of positive results, particularly because, in most cases, the detected nucleic acid does not indicate viable virus. Notably, positive results do not exclude bacterial or viral co-infection. False-positive influenza PCR results were reported associated with the administration of the live attenuated influenza vaccine (67).
False-positive molecular results may be due to an assay error or a contamination event (68). Usually, the most common sources of contamination are amplicon carryover or sample to sample cross-contamination (69). For conventional molecular tests with separate extraction and amplification steps, a unidirectional work flow that separates the clean (pre-amplification) from dirty (post-amplification) areas is an ideal layout that markedly reduces the chances of contamination. For sample-to-answer platforms and POCTs, amplicons contamination is less likely. In all cases, thorough cleaning and decontamination before and after any procedure reduce contamination. Cleaning using freshly made 10% to 15% sodium hypochlorite solution followed by 70% ethanol is recommended to eliminate or reduce contamination from amplified nucleic acids on laboratory surfaces. UV irradiation may be use depending on institutional guideline and with proper maintenance of the UV bulbs. Swipe testing performed at predefined schedules to screen surfaces that are liable to contamination is a good approach for identifying contamination events.
Generally, a single negative molecular result has a high negative predictive value, meaning that if a sample returns a negative result, the result is unlikely to be a false negative. However, false-negative molecular results have been reported and repeat molecular testing is recommended in certain situations when infection is highly suspected, or the adequacy of specimen collection is questionable. False-negative PCR results were reported in COVID-19 patients with symptoms, typical chest imaging, or seroconversion and varied by the duration of illness (70, 71). A false-negative influenza molecular test in influenza A-infected patients was also reported (72) and the CDC recommends that if no other etiologies are identified in hospitalized patients or those with LRTIs, repeat testing should be performed and antiviral treatment should be started (73). The kinetics of viral shedding and the duration of disease could contribute to false-negative results. Additional analytical causes of false negatives include target degradation due to improperly collected or stored specimens, viral evolution that can cause mutations in the primer or probe binding sites (e.g., novel influenza A strain or SARS-CoV-2 variants), and interference caused by inhibitors present in the clinical specimens.
Interpreting Detection of Multiple Viruses on Respiratory Panels
When testing with respiratory viral panels, codetection of several viral targets (e.g., greater than 4 viruses) in the same specimen is infrequent. The rates of co-infections vary widely with co-infections with SARS-CoV-2 being less common, while coinfections with rhinovirus/enteroviruses occurring more frequently (74–79). Co-infections with respiratory viruses is plausible, especially in children younger than 5 years (80), and are associated with unfavorable outcomes (81, 82). However, the medical significance of detecting multiple viruses is not well understood. Controlled studies that could differentiate the signatures and clinical significance of co-infections vs co-detections are needed (83). It is essential to carefully interpret the results of respiratory viral panels in light of the clinical presentation and crucial to work closely with the clinical diagnostic laboratories to understand the analytical performance of the assays and results’ interpretations.
Interpreting Antigen Testing
A positive antigen result can indicate an infection with the tested virus; however, a negative antigen test result does not rule out the infection given the lower sensitivity of antigen tests. Antigen testing is beneficial when molecular testing is not available or as a point-of-care or at-home test. Extensive evaluation of SARS-CoV-2 antigen testing showed high analytical specificity but lower analytical sensitivity when compared to molecular testing. Antigen testing was most sensitive when used for symptomatic patients within the first seven days of illness (84–87), and higher antigen sensitivity correlated with higher viral load in clinical samples. Considering the analytical performance of antigen testing, a positive result can indicate SARS-CoV-2 infection, and a negative result warrants repeat testing with a molecular method in symptomatic patients (88). With the progress of the COVID-19 pandemic, some reports suggested the utility of antigen testing for making decisions on patient isolation, based on correlations of antigen results and recovery of infectious virus on cell culture (89, 90). However, this approach had several limitations including the lower sensitivity of both antigen testing and cell culture (91, 92). In 2017, the FDA required minimum criteria of 80% sensitivity and 95% specificity compared to RT-PCR for approving influenza antigen testing for clinical use. Due to the lower sensitivity of influenza antigen testing, negative results should not exclude influenza infection in patients with clinical presentation consistent with influenza, particularly when the community transmission level is high. Generally, molecular detection of influenza is favored by the CDC and Infectious Disease Society of America (IDSA) for both inpatients and outpatients (93, 94). The sensitivity of RSV antigen testing was higher in children than adults, which limits its utility for diagnosis in elderly populations (95). Antigen testing for other respiratory viruses is less frequent and the reported sensitivity was variable, but generally lower than molecular-based methods.
Interpreting Correlates of Viral Loads
The correlation between viral load, level of infectivity, and the severity of respiratory disease has been an area of investigation. The Ct is the amplification cycle, when using real-time PCR, at which the fluorescence generated by target amplification crosses the threshold of the assay. A target with higher load will associate with an earlier Ct value. Of note, not all commercially available molecular tests provide Ct value information. The Ct values provided by qualitative SARS-CoV-2 assays were often utilized to indicate the relative viral RNA abundance and to guide infection control and patient management and were either reported clinically or communicated as “research use only” depending on the laboratories (96–98). Data on correlating Ct values with COVID-19 severity and prognosis were variable, likely due to the lack of standardization of the utilized methods, sample types, and the severity scoring approach (99, 100). The utility of Ct values during the COVID-19 pandemic included primarily guiding decisions on patient isolation, especially in asymptomatic, fully vaccinated, or previously infected patients. The unvalidated use of Ct values should be guided by the clinical scenario, history of exposure, vaccination status, and the limitations of the research use only of Ct values should be clearly communicated. Ct values should be generated by the same assay using the same sample type if they are intended to be compared to historical results. Given the limitations and variability associated with this marker, it is recommended that clinicians consult with their clinical microbiology laboratory director to clarify the analytical interpretations of Ct values. Also, for respiratory viruses other than SARS-CoV-2, the utility of quantification was proposed. Studies correlated viral load and invasive adenovirus disease, severity of influenza, RSV, and enterovirus (101–105). The clinical significance of respiratory viral quantification is uncertain and studies that utilize standardized quantitative methods are needed for understanding the utility of viral loads and their role in guiding patient management.
The detection of sub-genomic RNA (sgRNA), produced by discontinuous transcription of the SARS-CoV-2 genome (106), was a proposed approach for discriminating genomic RNA from actively replicating virus (107–109). A leader sequence transcribed with the target gene facilitates the differentiation between genomic and sub-genomic RNA (110). In a study that looked at the dynamics of detection of genomic and sgRNA, only 40% of samples had detectable sgRNA. At a viral load of 5.1 log10 copies/mL, 96% of the samples had detectable sgRNA, which was detected for a shorter duration than the genomic RNA (110). Currently, no assays are commercially available for the detection of SARS-CoV-2 sgRNA.
Recommendations
- Viral load test results should be interpreted in light of clinical symptoms.
- Positive molecular or antigen test results in patients without symptoms may reflect asymptomatic carriage, pre-symptomatic infection, or shedding following resolved infections. While shedding will be detected for longer by molecular tests, it is expected that antigen tests may also remain positive once the acute infection has resolved. A disclaimer to that effect may be added to a test report to highlight that point.
- Negative test results in symptomatic patients may be false-negative results and repeat testing is recommended. A disclaimer to that effect may be added to a test report to highlight that point.
- Unusual positivity rates that are discordant from local prevalence should be investigated.
- Co-infections with multiple viruses can occur but results with greater than 4 viruses are unusual and should be investigated.
- Correlates of viral loads (e.g., threshold cycle) provided by some NAATs should be interpreted with caution given the lack of standardization.

Fig. 1. Reasons for respiratory viruses testing. Summary of the different reasons for performing testing for respiratory viruses including diagnostic, surveillance, resistance, and serological testing.
WHY SHOULD TESTING BE PERFORMED TO DETECT RESPIRATORY VIRUSES?
The decision to test patients for respiratory viruses depends on several factors including (a) treatment and management of the patient in a timely manner (i.e., diagnostic testing), (b) identification of cases during an outbreak and for infection control in hospitalized patients (i.e., surveillance testing), (c) investigation of treatment failure (i.e., antiviral resistance testing), and (d) identification of immune responses (i.e., antibody testing) (Fig. 1).
Different tests and sample types may be used based on variables such as patient age, comorbidities, baseline immune status, and hospitalization as well as geography, season, and prevalence of the circulating respiratory virus in the community (Fig. 1). Most respiratory viruses demonstrate seasonal variations in prevalence, particularly in temperate areas (Table 1). Often, laboratories restrict testing for specific respiratory viruses during certain seasons. On the other hand, since NAATs and viral culture are highly specific, the positive predictive value of these tests remains high even during times of low viral prevalence.
Diagnostic Testing
Accurate and timely diagnosis of the viral cause of respiratory infections has several potential benefits including initiation of antivirals and discontinuation of antibiotics, decreasing the overall costs of care, reducing selection for antimicrobial resistance due to excessive antibiotic use, and improved patient and physician satisfaction (111). The literature supporting this is mixed, possibly due to differences in the patient populations, assay, or sample types employed.
Initiation of antiviral therapy. Currently, FDA-approved antiviral therapeutic agents for respiratory viral infections are restricted to the treatment of influenza and SARS-CoV-2. Diagnostic testing for influenza should be performed if the results of the test will influence subsequent initiation of antiviral therapy or prophylaxis for high-risk contacts (112). Outpatients with higher risk for influenza complications should be considered for testing if they are presenting within 48 h of symptom onset (112). Testing beyond this window may not be useful because the yield would be low, and the patient would be outside the window for antiviral therapy. Hospitalized patients and nursing home residents with febrile illness during influenza epidemics should also be tested, regardless of the duration of symptoms, to determine the potential for nosocomial transmission (113). While patients may be treated empirically when influenza is clinically suspected, a positive test result is most likely to prompt an intervention for influenza.
A systematic review of influenza POCTs vs higher complexity laboratory tests (e.g., NAAT, culture) or clinical diagnosis in ambulatory care settings showed that POCTs had no effect on admissions, returning for care, or the frequency of prescription of antibacterials, but found increased prescribing of antivirals (114). Further testing was reduced for full blood counts, blood cultures, and chest radiography. Time in the ED was not changed. On the other hand, a randomized clinical trial (115) among adults with acute respiratory illness admitted to 2 UK hospitals showed routine molecular point-of-care testing for influenza was associated with improved influenza detection and improvements in appropriate and timely antiviral and isolation facility use, due to the test’s accuracy, ease of use, and faster TAT. The utility of testing adult outpatients for viruses other than influenza and SARS-CoV-2 has been questionable. Adult outpatient outcomes were assessed at a Connecticut Veterans Affairs Center that used an on-demand respiratory panel (116). Outpatients were divided into those with influenza virus detected, those with a non-influenza virus detected, and those with no pathogen detected. The influenza-positive cohort was more likely to be treated with an antiviral agent compared to patients in the other two cohorts. Given the poor predictive value of the US CDC’s influenza-like illness criteria in adult transplant patients, routine multiplex respiratory NAAT assays are recommended in transplant patients with suspected respiratory virus infection for better diagnosis and improved antiviral use (117). Due to the clinical severity of illness, patients admitted to the intensive care unit (118) and pediatric patients with an underlying illness (119) may be good candidates for respiratory virus panel testing.
Discontinuation of antibiotics. For testing to impact decision-making, it is essential that the results be available within 24 h of sample collection (120). Batch testing performed 2 to 3 times a week is not optimal. A study including an adult population presenting to the ED with respiratory symptoms compared the use of a respiratory viral panel with an average TAT of 28 h with that of a respiratory pathogen panel with an average TAT of 3 h (121). Switching to a multiplex respiratory panel with a clinically actionable TAT was associated with reduced hospital admissions, and for admitted adults without focal radiographic findings, reduced antibiotic initiation (121). Another study showed that the FilmArray rapid respiratory panel decreased the duration of antibiotic use, the length of inpatient stay, and the time to isolation for children admitted to the hospital with an acute respiratory tract illness, compared to batched PCR analysis for RSV, and influenzas A and B, with additional testing for parainfluenza and hMPV in certain patients (122). Another study demonstrated that a significant decrease in admission rates, shorter lengths of stay, shorter durations of antimicrobial therapy, and fewer chest radiographs were observed in an adult tertiary care center when rapid panels were used compared to conventional antigen detection and older molecular methods (123).
The impact of rapid respiratory viruses testing on antibiotic use also varies. A study based in a large UK hospital showed that a routine molecular POCT for respiratory viruses in adults presenting with acute respiratory illness was associated with a reduction in length of stay, duration of antibiotics, and improved influenza detection and antiviral use. However, it did not reduce the proportion of patients treated with antibiotics (124). A randomized clinical trial among children with influenza-like illness in the ED (125) found that children with rapid respiratory pathogen test results were more likely to receive antivirals and be hospitalized; however, there was no significant difference in antibiotic prescribing. A similar trial showed that rapid respiratory pathogen testing was associated with a trend toward decreased antibiotic use, but it did not meet statistical significance (126). Another randomized clinical trial among 1243 children presenting to a pediatric ED with fever and respiratory symptoms (127) found that multiplex POCTs for respiratory pathogens did not reduce the use of antibiotics, number of unnecessary diagnostic tests, or costs. The lack of difference in the rates of antibiotic utilization with respiratory viral testing may be due to a sicker cohort of patients tested in these studies. Many of these patients required admission to the hospital and had clinical evidence of pneumonia, which required prolonged use of antibiotics.
Surveillance Testing
Hospital surveillance testing should usually be restricted to symptomatic patients who are epidemiologically linked to suspected outbreaks of a viral pathogen. Examples include travelers returning from a region with a high prevalence of a respiratory virus or individuals exposed during institutional outbreaks (113). Respiratory panel use allows for more comprehensive characterization of viruses for general epidemiology/surveillance and outbreak investigation. A fast TAT is equally important for surveillance purposes for efficient bed utilization and appropriate infection control measures, such as cohorting, to reduce nosocomial transmission, and to identify outbreak situations (120). Nosocomial viral infections impose a substantial burden in hospitals and pose a particular risk to immunocompromised hosts, particularly children and adults who receive solid organ transplant (SOT) and hematopoietic stem cell transplant (HSCT) (128). Improved molecular detection has led to increased recognition of hMPV, coronavirus, bocavirus, and rhinovirus infections in transplant recipients, prompting effective virus-specific cohorting and isolation, thereby curbing outbreaks in transplant units with horizontal transmission (111). In high-risk congregate settings, repeat screening of outbreaks, including SARS-CoV-2, may be useful in identifying individuals with viral infection, thereby enabling isolation. Identifying respiratory viruses can provide epidemiologic tracking of regional, national, and international outbreaks (129).
Genetic detection, identification, and characterization of novel infectious agents using NGS has been proven to be a powerful tool allowing for the identification of nosocomial and community outbreaks and characterizing circulating variants, and for the discovery of new routes of viral transmission (130). Several groups have published examples of the utility of NGS for respiratory viral outbreak investigation. Whole genome sequencing (WGS) data have been used to inform the real-time infection prevention response to a cluster of hospital-acquired human parainfluenza 3 virus infections at a children’s hospital (131). Similarly, partial and WGS assays demonstrated 100% identity across the entire adenoviral genome for cases of a neonatal intensive care unit adenoviral outbreak and samples taken from ophthalmologic equipment, prompting significant procedure changes for ophthalmologic equipment use and cleaning (132).
Antiviral Resistance Testing
Unlike bacterial infections, antimicrobial resistance testing is not routinely performed for viral infections and is primarily prompted by treatment failure as observed in symptomatic patients shedding viruses for prolonged periods. This is primarily true for influenza viruses. During the 2008 to 2009 influenza season, high rates of oseltamivir resistance (>90%) were observed in the United States and during the 2009 influenza A virus H1N1 pandemic, 37 cases of oseltamivir resistance were identified in the United States. Three-quarters of these cases occurred in immunocompromised patients, and 89% occurred in patients who had received oseltamivir (133). Since then, 99% of influenza virus isolates tested in the United States have been susceptible to neuraminidase inhibitors and there has been less need for resistance testing. Most clinical laboratories do not perform resistance testing on-site and, when testing is needed, samples are forwarded to public health or reference laboratories. NGS is now the method of choice to simultaneously provide strain typing and resistance testing, which in turn could be used to inform public health of circulating strains and allow rapid detection of the emergence of novel subtypes or highlight potential outbreaks (134).
Serological Testing
While not recommended for routine diagnosis of acute respiratory infections, serologic tests may be considered for the retrospective diagnosis of respiratory viral infections due to influenzas A and B, SARS-CoV-2, RSV, adenoviruses, and PIVs, particularly in seroprevalence surveys. However, serological testing has its own limitations. For example, IgM and IgG are not detectable in patients acutely infected with COVID-19 as it may take days to weeks after the infection for antibodies to become detectable. Additionally, antibody testing is not recommended to determine response to vaccination as precise immune correlates of protection remain uncertain. In a meta-analysis of 29 studies that evaluated the diagnostic accuracy of IgG- and IgM-based POCTs that detect SARS-CoV-2 antigens, a combined IgG/IgM test had better sensitivity than measuring either antibody type independently (135). In another systematic review of 38 studies that evaluated the sensitivity of serologic testing by time from symptom onset in patients with COVID-19, IgM was detected in 23% by 1 week, in 58% by 2 weeks, and in 75% by 3 weeks; the corresponding detection rates for IgG were 30%, 66%, and 88% (136). This is true for novel viral infections such as SARS-CoV-2, which did not circulate prior to December 2019. In contrast, for infections such as influenza which have been circulating for several decades, many individuals would have positive IgG in the setting of prior exposure. Hence, improving sensitivity for IgG would be at the expense of specificity.
Recommendations
- Testing should be performed if there is high pretest probability of respiratory viral infection based on clinical presentation and local prevalence.
- Testing should be performed in the following scenarios (a) if the results will change management (for example, initiation of appropriate antivirals, discontinuation of unnecessary antibiotics), (b) infection control guidance (for example, implementation of appropriate isolation measures, cohorting of patients, and surveillance during outbreak situations), or (c) evaluation of local seroprevalence.
WHO SHOULD BE TESTED TO DETECT RESPIRATORY VIRUSES?
The goal of efficient respiratory virus testing is to optimize a strategy that leads to the best patient outcomes. Factors such as time to result availability, clarity of diagnosis, and ability to guide treatment and patient isolation and cohorting decisions are key parts of developing the best testing strategy, but the significance of each one of these factors varies across different patient populations and can also be highly dependent upon which respiratory viral pathogen is detected. In the hospitalized inpatient and immunocompromised populations, all of these factors are essential, while in the outpatient setting, in individuals that are otherwise healthy, testing, when warranted, is typically focused on a targeted number of respiratory pathogens, such as influenza and SARSCoV-2 (Fig. 2).
Testing in the Pediatric Population
It is estimated that preschool-age children can have 6 to 10 viral colds a year (137, 138). In addition, one-third of pediatric primary consultations are due to acute respiratory infections in the United Kingdom (138). In the pediatric population, the respiratory pathogens most commonly identified are rhinovirus/enterovirus, RSV, influenza virus, adenovirus, PIVs, and coronaviruses (139). While the clinical utility of viral testing in the pediatric population is questioned in certain contexts, the general concerns raised focus on whether testing results will be properly utilized by the ordering clinician to drive clinical decision-making, specifically in the healthy and immune-competent children in the outpatient setting (32, 140). In children who are hospitalized, or have underlying health conditions, the case for respiratory virus testing is more clearly demonstrated, with multiple studies showing the clinical utility of testing for both treatment decisions and infection control purposes (141–143).
While there was a lower rate of mortality and morbidity due to SARS-CoV-2 observed in children compared to older adults, severe disease has been reported in children. In addition, there have been reports that children play a role in the transmission of COVID-19 (144). During SARSCoV- 2 infection, it was shown that asymptomatic children may have significantly higher viral loads than symptomatic children, pointing out the importance of masking, social distancing, and testing strategies to limit the spread of infection (145).
Testing in the Aging Population and Immunocompromised Hosts
In the aging population and in immunocompromised patients, respiratory viral infections lead to significant morbidity and mortality underscoring the need for rapid testing to influence patient management. A pre-COVID-19 pandemic study showed a significant increase in acute respiratory infections requiring hospitalization in elderly patients over a 10-year period (146). Influenza, RSV, and recently SARS-CoV-2 are all major causes of morbidity and mortality in the elderly and immunocompromised populations. In addition, other outbreaks, such as hMPV, have also caused severe viral infections in long-term care facilities for the elderly (147). Hospitalization rates and the overall risk of death have increased in those 65 years of age and older for influenza infections (148), with 50% to 70% of influenza- related deaths occurring in this age group (7, 149, 150). Viral pneumonia is 10 times more likely in the elderly population than in younger adults (151) and leads to worse outcomes (152, 153).
A recent literature review estimated 1.5 million episodes of RSV in adults 65 years of age and older in industrialized countries in 2015, with approximately 14.5% of these cases requiring a hospital admission (154). RSV infection is also a substantial cause of morbidity and mortality in immunocompromised populations, with RSV infections in HSCT and SOT ranging from 1% to 12%. An estimated 18% to 55% of these infections progress to LRTIs, and death occurs in 7% to 33% of cases (155, 156). The detection of RSV in the pretransplant period can result in postponement of transplant given the higher risk for increased morbidity and mortality (18) In one study of HSCT recipients, 80% to 90% of patients with upper respiratory RSV infection developed pneumonia with 30% to 40% exhibiting symptoms within 7 days following URTI symptoms (30).
In some studies, for oncology patients diagnosed with influenza, parainfluenza, RSV and adenovirus, antivirals when available and administered early have been shown to improve outcomes (157, 158). Therefore, testing for influenza, RSV, parainfluenza, and other prevalent community-circulating viruses in all symptomatic patients with malignancies is recommended since these results may be used to guide antiviral treatment decisions. Similarly, transplant recipients may also be an appropriate patient population for multiplex testing (159).
Testing in the Immunocompetent Adult Population
Inpatient setting. In the inpatient population, the clinical utility of testing for viral respiratory pathogens is important for guiding treatment and infection control decisions. It is also clear from the literature that cases of respiratory infections are generally underestimated. One study looking at a period from 2003 to 2014 found that a quarter of influenza-attributable hospitalizations did not have a definitive acute respiratory infection diagnosis (160). Another study performed in the United States estimated that only 1 in 10 influenza-associated critical illnesses in intensive care unit patients included a formal diagnosis of influenza (161).
For influenza, rapid molecular test results have led to earlier initiation of appropriate therapy (116, 162, 163). Additional respiratory testing in the inpatient population using small, targeted panels (e.g., Flu/RSV, Flu/RSV/SARS-CoV-2) and rapid multiplex respiratory panels has also been shown to positively impact certain patient outcomes (121–123). Another study showed that the use of a multiplex pneumonia panel in critically ill COVID-19 patients with suspected bacterial respiratory superinfection led to a de-escalation of empiric antibiotic therapy in two-thirds of patients and also allowed the clinical team to prevent initiation of empiric therapy in two-thirds of cases (164).
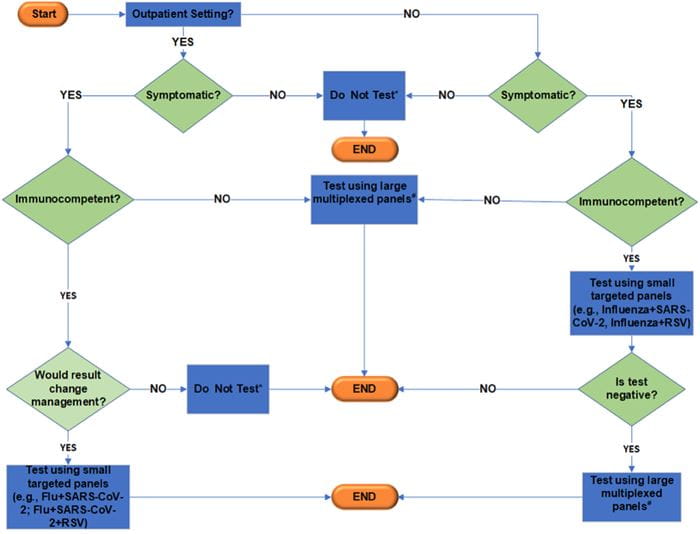
Fig. 2. Suggested testing algorithm for respiratory viruses. *Testing might be necessary for certain situations (e.g., surveillance screening or outbreak investigations). Additional considerations include symptoms onset, age of patients, sample types, and test availability. #Multiplexed panels (panels with additional targets beyond Flu/RSV/SARS-CoV-2).
Outpatient setting. In the outpatient setting, viruses are the most common pathogens identified in severe cases of community-acquired respiratory infections (151). It is also clear that in the outpatient population, the burden of respiratory viral infection is also significantly underestimated. The CDC published testing guidance for when SARS-CoV-2 and influenza viruses are co-circulating. In cases where a patient presents to the outpatient clinic or the ED with symptoms of acute respiratory illness that require hospitalization, the recommendation is to minimally test for influenza A, influenza B, and SARS-CoV-2. If the patient does not require hospitalization, the recommendation is to test for SARS-CoV-2 and also test for influenza if the result will have an impact on clinical management or infection control, or treat empirically (165). In immunocompetent adult patients, RSV causes a mild illness that often resolves within 5 days. For patients presenting in an outpatient setting with more severe diseases or with underlying chronic infections, testing for RSV may be warranted, similar to recommendations for immunocompromised patients (166).
Recommendations
- Testing of pediatric patients should be limited to hospitalized children or children with underlying conditions. Depending on the season, small, targeted panels (e.g., influenza/RSV) might be sufficient.
- Aging, ill patients, and immunocompromised patients should be tested using multiplexed respiratory pathogens panels.
- Immunocompetent adult patients should be tested if results will impact management, primarily for influenza and SARS-CoV-2.
WHAT IS THE ROLE OF DIAGNOSTIC STEWARDSHIP IN RESPIRATORY VIRUS TESTING?
Diagnostic stewardship aims to select the right test for the right patient, generating accurate, clinically relevant results at the right time to guide appropriate clinical behavior, while conserving healthcare resources (167). Selection of the right test involves the evaluation of test performance, testing volume, laboratory feasibility, cost vs value, and the overall impact on clinical outcomes. Provider education efforts and guidance in the form of algorithms and policies can be useful tools to guide appropriate test selection (e.g., Fig. 2). For instance, in a healthcare system based in Indiana, an algorithm was developed for patients presenting to the ED and hospital clinics with influenza-like illnesses to guide providers to select appropriate assay. Implementation of this protocol resulted in improvement in the appropriateness of ordering and significant cost savings (168). In another study, a hospital-wide diagnostic policy for implementation of rapid diagnostic assay for the detection of respiratory viruses in patients presenting to the ED was developed. Nearly half of the patients who tested positive for a respiratory virus did not receive antibiotics. The value-based measure, expressed in euro-hour, increased 10-fold compared to their former policy (169).
The fixed nature of the multiplex PCR panels raises the concern that they might include pathogens that the clinician does not want to test for. Ideally, common pathogens should be tested first followed by testing for uncommon pathogens. For instance, infection with Chlamydophila pneumoniae is uncommon enough that routine diagnostic testing is not recommended for community-acquired pneumonia in adults (170). However, several multiplex respiratory pathogen panels test for Chlamydophila pneumoniae in combination with testing for other common respiratory viruses. Inappropriate use of tests in cases with low pretest probability can lead to misinformation from false-positive results, potentially misleading clinicians and adding to healthcare costs. Currently, these choices are limited by lack of commercially available pathogen-specific PCR tests.
Besides influenza, RSV, and SARS-CoV-2 NAATs, clinicians are currently unable to order targeted PCR for any other respiratory viral pathogen except possibly using LDTs; hence, there is no choice left but to order the multiplex PCR panels. To prevent overutilization of rapid multiplex viral panels by clinicians in low-impact situations, indication selection using restrictive or guided test ordering built into the electronic medical records can be used. This requires clinicians to input an approved indication from a list of appropriate use criteria before ordering the rapid multiplex viral panel. If patients do not meet appropriate use criteria, approval from infectious disease providers or clinical laboratory directors may be indicated.
Recommendations
- Educational material should be available to clinicians to guide respiratory tests selection.
- Built-in electronic medical record algorithms to drive appropriate test selection and ordering should be considered.
- In general, small, multiplexed panels or targeted NAATs should be used first when available instead of broad multiplexed panels unless patients are immunocompromised.
WHAT ARE THE RECOMMENDATIONS FROM OTHER PROFESSIONAL SOCIETIES?
This section is a high-level summary covering some key aspects of guidelines from other professional societies. For the laboratory detection of respiratory viruses, both the American Society of Microbiology (ASM) and the IDSA recommend molecular testing as the reference method (83, 171).
Published guidelines by the ASM for current best practices in respiratory virus testing point out that the use of NAATs for detection of respiratory viruses is superior to traditional methods, such as culture, DFA, or rapid antigens tests, due to their increased sensitivity and specificity (171). The importance of being aware of viral circulation patterns and ensuring that laboratory testing matches these patterns is also highlighted (e.g., by consulting CDC and other public health databases), as well as the criticality of good laboratory practices to avoid contamination with the use of molecular assays due to their increased sensitivity. Consideration of the patient’s immune status is also addressed, pointing out the importance of testing in immunocompromised patients vs immunocompetent patients (171). Additional guidelines published by the ASM for multiplex panels, including respiratory pathogen panels, highlight the benefits of these panels, which include diagnostic performance and time to results, and an increasing number of studies show the benefits in both patient care and healthcare cost reduction (172).
For hospitalized adult patients, the IDSA recommends that when influenza is known to be circulating, all patients with acute respiratory symptoms (upon admission or developing during hospitalization) or acute worsening of chronic cardiopulmonary disease, should be tested. Influenza testing during periods of low viral activity is recommended for patients that present with acute respiratory illness and have a known exposure to someone with influenza or another respiratory illness of unknown cause or have been to an area with influenza activity. In addition, testing during a low incidence of influenza activity is also recommended for patients with an acute respiratory illness, especially immunocompromised adults and children, if it will influence prophylaxis treatment of high-risk members of the same household (112). For influenza testing in the outpatient setting, the IDSA recommends that when seasonal influenza is circulating in the local community, high-risk patients and those with chronic medical conditions should be tested for influenza if it will influence clinical management. In addition, it is recommended that symptomatic patients not at high risk for complications should be tested if the result will impact treatment decisions or may influence treatment decisions for high-risk members of the same household (112). In addition, the IDSA clinical practice guidelines for seasonal influenza suggest that even during periods of low influenza activity, clinicians can consider influenza testing in patients with acute respiratory symptom onset, especially in immunocompromised and high-risk patients in the outpatient setting (112). With varying levels of evidence, the IDSA recommends testing of symptomatic individuals for SARS-CoV-2, even if there is a low clinical suspicion. They also recommend testing of asymptomatic individuals in several different scenarios, such as COVID-19 exposures and before certain medical procedures (173).
With regards to influenza, the American Academy of Pediatrics (AAP) recommends that, “Testing should be performed when timely results will be available to influence clinical management or infection-control measures” (174). In addition, the most recent guideline updates from the AAP recommends SARS-CoV-2 testing for any children who have symptoms of COVID-19, have had close contact with a confirmed or probable case of COVID-19, or those who are required to have screening testing (175). The AAP has also updated guidance following a surge in RSV cases in the spring of 2021, pointing out the need to have adequate diagnostic testing, especially in children who are at high risk (175).
The American Thoracic Society (ATS) guidelines (176) recommend against performing nucleic-based testing of respiratory samples for viral pathogens other than influenza for outpatients with suspected community-acquired pneumonia (CAP); however, for hospitalized patients with severe CAP or for immunocompromised patients, nucleic-based testing is suggested. The Infectious Diseases Working Party of the German Society for Hematology and Medical Oncology identified community-acquired respiratory virus infection as a significant cause of morbidity and mortality in patients with underlying malignancy (177).
Per IDSA recommendations, serologic testing should not be used to diagnose SARS-CoV-2 infection during the first 2 weeks following symptom onset. Due to the relatively higher accuracy of SARS-CoV-2 IgG antibody or total antibody tests compared to IgM antibody, IgA antibody, or IgM/IgG differentiation tests, SARS-CoV-2 IgG antibody or total antibody are the preferred diagnostic tests to identify patients who previously had SARS-CoV-2 infection, as well as patients with current infection who have had symptoms for 3 to 4 weeks (178).
FUTURE CONSIDERATIONS
The Laboratory Response Network (LRN) was established by the CDC to respond to public health threats, including novel respiratory pathogens. The network includes local and state public health laboratories, federal, military, veterinary, and international laboratories. The LRN laboratories receive appropriate training in addition to standardized reagents and protocols for testing clinical specimens with a potential novel pathogen under investigation. The reference laboratories (i.e., LRN member laboratories) can then train sentinel laboratories. Sentinel laboratories are essential for early identification and referral to LRN laboratories for further testing. The FDA can issue an EUA for unapproved diagnostics to facilitate the availability of diagnostics in situations of public health emergencies. The first EUA test allowed the development of an assay for diagnosing influenza A(H1N1) pdm09 during the 2009 influenza pandemic. This venue enabled nationwide implementation and increased testing capacity for SARS-CoV-2 with the beginning of the COVID-19 pandemic, yet after an initial delay in allowing the clinical microbiology laboratories and industry to offer testing and independently apply for EUA approvals (179). With this infrastructure, implementing diagnostic assays for a novel pathogen remains a challenge. The availability of control materials and clinical specimens required for the assays’ evaluation and the lack of gold standard reference methods to assess the clinical sensitivity and specificity are the most challenging. The speed in implementing diagnostics can have a significant value in limiting the disease transmission. Regulations, intended to ensure the safety and performance of diagnostic assays, might slow down early diagnosis and disease containment. In situations when prompt and large-scale implementation of diagnostics become required via high-throughput and point-of-care testing, this could be challenged by assays’ development and safety regulations. Even though the EUA mechanism allows for a rapid response to an emergent respiratory pathogen, the COVID-19 pandemic emphasized the significance of the quick large-scale access to diagnostics, which was successfully applied during the monkeypox outbreak (180). The close and prompt collaboration between the FDA, CDC, LRN, and academic and reference laboratories to quickly implement and expand testing utilizing the most optimized methods should enhance an early response to outbreaks of novel pathogens.
CONCLUSIONS
The recent COVID-19 pandemic and prior to that the 2009 H1N1 influenza pandemic highlight the need for laboratories to be alert and prepared for the diagnosis of emerging respiratory infections. It is important for laboratories to monitor patterns of detection of respiratory pathogens targeted by current methods for unusual results that may suggest an emerging or changing virus. For example, many of the respiratory multiplexed panels include detection and genotyping of influenza A virus. An increase in the detection of un-typable influenza A virus may suggest a drift or shift in the virus or the emergence of a novel influenza virus. Additionally, new therapies including the recent FDA-approved Pfizer RSV vaccine (ABRYSVO™) for older adults and pregnant individuals will likely impact the epidemiology of RSV in the future. As antiviral therapeutic options eventually expand beyond influenza and RSV, the need for rapid testing will expand and recommendations for testing will need to be revised accordingly (181).
One of the outcomes from the COVID-19 pandemic was the expansion of NGS methods including WGS and metagenomics, in both public health and clinical laboratories for the purpose of SARS-CoV-2 variant surveillance. Data were widely shared across the world through either the Global Initiative on Sharing All Influenza Data or NCBI Virus and allowed rapid identification and notification of emerging SARS-CoV-2 variants. As NGS becomes more established in clinical laboratories, a similar approach of viral sequencing and data sharing has the potential to provide rapid identification and characterization of novel viruses.
The COVID-19 pandemic also saw an increase in the number of platforms with more accessible testing options including POCTs and at-home testing, both antigen-based and molecular tests. Many of these platforms might eventually expand testing to other viruses including influenza and RSV and likely any other emerging viruses. Laboratorians should remain alert and involved to provide guidance on managing testing and the information obtained from a wider range of testing settings.
Nonstandard Abbreviations: RSV, respiratory syncytial virus; hMPV, human metapneumovirus; MERS-CoV, Middle East respiratory syndrome; EUA, Emergency Use Authorization; POCT, point-of-care test; ED, emergency department; NAAT, nucleic acid amplification test; DFA, direct fluorescent antibody; TAT, turnaround time; NGS, next-generation sequencing; Ct, threshold cycle; IDSA, Infectious Disease Society of America; sgRNA, sub-genomic RNA; LRN, Laboratory Response Network.
Author Contributions: The corresponding author takes full responsibility that all authors on this publication have met the following required criteria of eligibility for authorship: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved. Nobody who qualifies for authorship has been omitted from the list. Gregory J. Berry (Conceptualization-Equal, Writing—original draft-Equal, Writing—review & editing-Equal), Tulip A. Jhaveri (Conceptualization-Equal, Writing—original draft-Equal, Writing—review & editing-Equal), Paige M.K. Larkin (Conceptualization- Equal, Writing—original draft-Equal, Writing—review & editing-Equal), Heba Mostafa (Conceptualization-Equal, Writing—original draft-Equal, Writing—review & editing-Equal), and N. Esther Babady (Conceptualization-Lead, Project administration-Lead, Writing —original draft-Lead, Writing—review & editing-Lead)
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form.
Research Funding: Funded in part through the NIH/NCI Cancer Center Support Grant (P30 [CA008748]).
Disclosures: N.E. Babady, grants and consulting fees from Roche/GenMark, ArcBio, Bio-Rad and Copan; honoraria from Roche; Evidence Based Laboratory Guidelines Subcommittee Chair for American Society for Microbiology and President of Pan-American Society for Clinical Virology. P.M.K. Larkin, grants from Abbott and BD, employment at ASM. T.A. Jhaveri, American Society of Microbiology (ASM)—invited presentation on “Top Papers in Diagnostic Stewardship” at ASM Microbe, 2022 (reimbursement for the symposium and related travel). H. Mostafa, grants from CIERS HHSN272201400007C, The Johns Hopkins University, Qiagen, Bio-Rad, and Hologic; consulting fees from BD Diagnostics and Seegene; honoraria from BD Diagnostics and Bio-Rad; support for attending meetings and/or travel from Bio-Rad. G.J. Berry, consulting fees from Cepheid, VedaBio, BioFire, Roche, Abbott, QuidelOrtho (formerly Quidel); honoraria from Cepheid.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
REFERENCES
- Hawkes MT, Lee BE, Kanji JN, Zelyas N, Wong K, Barton M, et al. Seasonality of respiratory viruses at northern latitudes. JAMA Netw Open 2021;4:e2124650.
- CDC. National Hospital Ambulatory Medical Care Survey: 2019 emergency department summary tables. 2019. https://www.cdc.gov/nchs/data/nhamcs/ web_tables/2019-nhamcs-ed-web-tables-508.pdf (Accessed February 2024).
- Hall CB. The spread of influenza and other respiratory viruses: complexities and conjectures. Clin Infect Dis 2007;45:353–9.
- Leung NHL. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol 2021;19:528–45.
- Ison M.G., Hirsch H.H. Community-acquired respiratory viruses in transplant patients: Diversity, impact, unmet clinical needs. Clin Microbiol Rev 2019;32:716–47.
- Mistry RD, Fischer JB, Prasad PA, Coffin SE, Alpern ER. Severe complications in influenza-like illnesses. Pediatrics 2014;134:e684–90.
- Zhou H, Thompson WW, Viboud CG, Ringholz CM, Cheng PY, Steiner C, et al. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008. Clin Infect Dis 2012;54: 1427–36.
- Barnes M, Heywood AE, Mahimbo A, Rahman B, Newall AT, Macintyre CR. Acute myocardial infarction and influenza: a meta-analysis of case-control studies. Heart 2015;101:1738–47.
- Dawood FS, Chaves SS, Perez A, Reingold A, Meek J, Farley MM, et al. Complications and associated bacterial coinfections among children hospitalized with seasonal or pandemic influenza, United States, 2003–2010. J Infect Dis 2014;209:686–94.
- Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev 2011;24:210–29.
- Newton AH, Cardani A, Braciale TJ. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol 2016;38:471–82.
- Turner RB. The common cold. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. 7th Ed. Philadelphia (PA): Churchill Livingston Elsevier; 2010. p. 809–13.
- Boyton RJ, Openshaw PJ. Pulmonary defences to acute respiratory infection. Br Med Bull 2002;61:1–12.
- Sieling WD, Goldman CR, Oberhardt M, Phillips M, Finelli L, Saiman L. Comparative incidence and burden of respiratory viruses associated with hospitalization in adults in New York City. Influenza Other Respir Viruses 2021;15:670–7.
- Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010;23:74–98.
- Srinivasan A, Wang C, Yang J, Inaba H, Shenep JL, Leung WH, Hayden RT. Parainfluenza virus infections in children with hematologic malignancies. Pediatr Infect Dis J 2011;30:855–9.
- Srinivasan A, Wang C, Yang J, Shenep JL, Leung WH, Hayden RT. Symptomatic parainfluenza virus infections in children undergoing hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2011;17: 1520–7.
- Cherukuri A, Patton K, Gasser RA Jr, Zuo F, Woo J, Esser MT, Tang RS. Adults 65 years old and older have reduced numbers of functional memory T cells to respiratory syncytial virus fusion protein. Clinical and vaccine immunology: CVI 2013;20:239–47.
- de Bree GJ, Heidema J, van Leeuwen EM, van Bleek GM, Jonkers RE, Jansen HM, et al. Respiratory syncytial virus-specific CD8+ memory T cell responses in elderly persons. J Infect Dis 2005;191:1710–8.
- Watson A, Wilkinson TMA. Respiratory viral infections in the elderly. Ther Adv Respir Dis 2021;15: 1753466621995050.
- Alcaide ML, Bisno AL. Pharyngitis and epiglottitis. Infect Dis Clin North Am 2007;21:449–69.
- Stolz D, Christ-Crain M, Gencay MM, Bingisser R, Huber PR, Muller B, Tamm M. Diagnostic value of signs, symptoms and laboratory values in lower respiratory tract infection. Swiss Med Wkly 2006;136:434–40.
- Donowitz GR. Acute pneumonia. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. 7th Ed. Philadelphia (PA): Churchill Livingston Elsevier; 2010. p. 891–916.
- Muller B, Harbarth S, Stolz D, Bingisser R, Mueller C, Leuppi J, et al. Diagnostic and prognostic accuracy of clinical and laboratory parameters in community-acquired pneumonia. BMC Infect Dis 2007;7:10.
- Spencer S, Thompson MG, Flannery B, Fry A. Comparison of respiratory specimen collection methods for detection of influenza virus infection by reverse transcription-PCR: a literature review. J Clin Microbiol 2019;57:e00027-19.
- Talbot HK, Falsey AR. The diagnosis of viral respiratory disease in older adults. Clin Infect Dis 2010;50:747–51.
- Loens K, Van Heirstraeten L, Malhotra-Kumar S, Goossens H, Ieven M. Optimal sampling sites and methods for detection of pathogens possibly causing community-acquired lower respiratory tract infections. J Clin Microbiol 2009;47:21–31.
- Treuhaft MW, Soukup JM, Sullivan BJ. Practical recommendations for the detection of pediatric respiratory syncytial virus infections. J Clin Microbiol 1985;22:270–3.
- Brendish NJ, Schiff HF, Clark TW. Point-of-care testing for respiratory viruses in adults: the current landscape and future potential. J Infect 2015;71:501–10.
- Azar MM, Landry ML. Detection of influenza A and B viruses and respiratory syncytial virus by use of clinical laboratory improvement amendments of 1988 (CLIA)-waived point-of-care assays: a paradigmshift to molecular tests. J Clin Microbiol 2018;56: e00367-18.
- Egilmezer E, Walker GJ, Bakthavathsalam P, Peterson JR, Gooding JJ, Rawlinson W, Stelzer-Braid S. Systematic review of the impact of point-of-care testing for influenza on the outcomes of patients with acute respiratory tract infection. Rev Med Virol 2018;28:e1995.
- Gill PJ, Richardson SE, Ostrow O, Friedman JN. Testing for respiratory viruses in children: to swab or not to swab. JAMA Pediatr 2017;171:798–804.
- van Elden LJ, van Kraaij MG, Nijhuis M, Hendriksen KA, Dekker AW, Rozenberg-Arska M, van Loon AM. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis 2002;34:177–83.
- Larkin PMK, Manuel V, Hernandez N, Garner OB. Novel use of rapid antigen influenza testing in the outpatient setting to provide an early warning sign of influenza activity in the emergency departments of an integrated health system. J Clin Microbiol 2020;58:e01560-20.
- Walter JM, Wunderink RG. Severe respiratory viral infections: new evidence and changing paradigms. Infect Dis Clin North Am 2017;31:455–74.
- Casiano-Colon AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol 2003;28:169–74.
- Allen KE, Chommanard C, Haynes AK, Erdman DD, Gerber SI, Kim L. Respiratory syncytial virus testing capabilities and practices among national respiratory and enteric virus surveillance system laboratories, United States, 2016. J Clin Virol 2018;107: 48–51.
- Ford L, Lee C, Pray IW, Cole D, Bigouette JP, Abedi GR, et al. Epidemiologic characteristics associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen-based test results, real-time reverse transcription polymerase chain reaction (rRT-PCR) cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clin Infect Dis 2021;73:e1348–55.
- Klajmon A, Olechowska-Jarzab A, Salamon D, Sroka-Oleksiak A, Brzychczy-Wloch M, Gosiewski T. Comparison of antigen tests and qPCR in rapid diagnostics of infections caused by SARS-CoV-2 virus. Viruses 2021;14:17.
- Navero-Castillejos J, Casals-Pascual C, Narvaez S, CuestaG, Hurtado JC, Fernandez M, et al. Diagnostic performance of six rapid antigen tests for SARS-CoV-2. Microbiol Spectr 2022;10:e0235121.
- Vemula SV, Zhao J, Liu J, Wang X, Biswas S, Hewlett I. Current approaches for diagnosis of influenza virus infections in humans. Viruses 2016;8:96.
- Kumar S, Henrickson KJ. Update on influenza diagnostics: lessons from the novel H1N1 influenza A pandemic. Clin Microbiol Rev 2012;25:344–61.
- Bakerman P, Balasuriya L, Fried O, Tellez D, Garcia-Filion P, Dalton H. Direct fluorescent-antibody testing followed by culture for diagnosis of 2009 H1N1 influenza A. J Clin Microbiol 2011;49:3673–4.
- Mandelia Y, Procop GW, Richter SS, Worley S, Liu W, Esper F. Optimal timing of repeat multiplex molecular testing for respiratory viruses. J Clin Microbiol 2020;58: e01203-19.
- Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015;28:465–522.
- WHO. Middle East respiratory syndrome coronavirus (MERS-CoV) https://www.who.int/health-topics/middleeast- respiratory-syndrome-coronavirus-mers#tab= tab_1 (Accessed November 2022).
- CDC. MERS in the U.S. https://www.cdc.gov/coronavirus/ mers/us.html (Accessed November 2022).
- Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 2019;14:319–38.
- Ong DSY, Fragkou PC, Schweitzer VA, Chemaly RF, Moschopoulos CD, Skevaki C, European Society of Clinical M, Infectious Diseases Study Group for Respiratory V. How to interpret and use COVID-19 serology and immunology tests. Clin Microbiol Infect 2021;27:981–6.
- Zhang YV, Wiencek J, Meng QH, Theel ES, Babic N, Sepiashvili L, et al. AACC practical recommendations for implementing and interpreting SARS-CoV-2 emergency use authorization and laboratory-developed test serologic testing in clinical laboratories. Clin Chem 2021; 67:1188–200.
- Drain PK. Rapid diagnostic testing for SARS-CoV-2. N Engl J Med 2022;386:264–72.
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 2020;382:2081–90.
- Suess T, Remschmidt C, Schink SB, Schweiger B, Heider A, Milde J, et al. Comparison of shedding characteristics of seasonal influenza virus (sub)types and influenza A(H1N1) pdm09; Germany, 2007–2011. PLoS One 2012;7:e51653.
- Loeb M, Singh PK, Fox J, Russell ML, Pabbaraju K, Zarra D, et al. Longitudinal study of influenza molecular viral shedding in Hutterite communities. J Infect Dis 2012; 206:1078–84.
- Birger R, Morita H, Comito D, Filip I, Galanti M, Lane B, et al. Asymptomatic shedding of respiratory virus among an ambulatory population across seasons. mSphere 2018;3:e00249-18.
- Gniazdowski V, Morris CP, Wohl S, Mehoke T, Ramakrishnan S, Thielen P, et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis 2020;73:e860–9.
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020;26:672–5.
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9.
- Gombar S, Chang M, Hogan CA, Zehnder J, Boyd S, Pinsky BA, Shah NH. Persistent detection of SARS-CoV-2 RNA in patients and healthcare workers with COVID-19. J Clin Virol 2020;129:104477.
- Fleury H, Burrel S, Balick Weber C, Hadrien R, Blanco P, Cazanave C, Dupon M. Prolonged shedding of influenza A(H1N1)v virus: two case reports from France 2009. Euro Surveill 2009;14:19434.
- Wang Y, Guo Q, Yan Z, Zhou D, Zhang W, Zhou S, et al. Factors associated with prolonged viral shedding in patients with avian influenza A(H7N9) virus infection. J Infect Dis 2018;217:1708–17.
- Zlateva KT, de Vries JJ, Coenjaerts FE, van Loon AM, Verheij T, Little P, et al. Prolonged shedding of rhinovirus and re-infection in adults with respiratory tract illness. Eur Respir J 2014;44:169–77.
- Morris CP, Luo CH, Sachithanandham J, Li M, Schwartz M, Gaston DC, et al. Large scale SARS-CoV-2 molecular testing and genomic surveillance reveal prolonged infections, protracted RNA shedding, and viral reinfections. Front Cell Infect Microbiol 2022;12: 809407.
- van der Vries E, Stittelaar KJ, van Amerongen G, Veldhuis Kroeze EJ, de Waal L, Fraaij PL, et al. Prolonged influenza virus shedding and emergence of antiviral resistance in immunocompromised patients and ferrets. PLoS Pathog 2013;9:e1003343.
- de Lima CR, Mirandolli TB, Carneiro LC, Tusset C, Romer CM, Andreolla HF, et al. Prolonged respiratory viral shedding in transplant patients. Transpl Infect Dis 2014; 16:165–9.
- 6Lehners N, Tabatabai J, Prifert C, Wedde M, Puthenparambil J, Weissbrich B, et al. Long-term shedding of influenza virus, parainfluenza virus, respiratory syncytial virus and nosocomial epidemiology in patients with hematological disorders. PLoS One 2016;11:e0148258.
- Curran T, McCaughey C, Ellis J, Mitchell SJ, Feeney SA, Watt AP, et al. False-positive PCR results linked to administration of seasonal influenza vaccine. J Med Microbiol 2012;61(Pt 3):332–8.
- Braunstein GD, Schwartz L, Hymel P, Fielding J. False positive results with SARS-CoV-2 RT-PCR tests and how to evaluate a RT-PCR-positive test for the possibility of a false positive result. J Occup Environ Med 2021;63:e159–62.
- Huggett JF, Benes V, Bustin SA, Garson JA, Harris K, Kammel M, et al. Cautionary note on contamination of reagents used for molecular detection of SARS-CoV-2. Clin Chem 2020;66:1369–72.
- Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020;173:262–7.
- Woloshin S, Patel N, Kesselheim AS. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med 2020;383:e38.
- Rello J, Rodríguez A, Ibañez P, Socias L, Cebrian J, Marques A, et al. Intensive care adult patients with severe respiratory failure caused by influenza A (H1N1)v in Spain. Crit Care 2009;13:R148.
- CDC. Information on Rapid Molecular Assays, RT-PCR, and other Molecular Assays for Diagnosis of Influenza Virus Infection. https://www.cdc.gov/flu/professionals/ diagnosis/molecular-assays.htm (Accessed February 2023).
- Eldesouki RE, Uhteg K, Mostafa HH. The circulation of non-SARS-CoV-2 respiratory viruses and coinfections with SARS-CoV-2 during the surge of the omicron variant. J Clin Virol 2022;153:105215.
- Uhteg K, Amadi A, Forman M, Mostafa HH. Circulation of non- SARS-CoV-2 respiratory pathogens and coinfection with SARS-CoV-2 amid the COVID-19 pandemic. Open Forum Infect Dis 2021;9:ofab618.
- Pretorius MA, Madhi SA, Cohen C, Naidoo D, Groome M, Moyes J, et al. Respiratory viral coinfections identified by a 10-plex real-time reverse-transcription polymerase chain reaction assay in patients hospitalized with severe acute respiratory illness–South Africa, 2009–2010. J Infect Dis 2012;206(Suppl 1):S159–65.
- Renois F, Talmud D, Huguenin A, Moutte L, Strady C, Cousson J, et al. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription-PCR DNA microarray systems. J Clin Microbiol 2010;48:3836–42.
- Canducci F, Debiaggi M, Sampaolo M, Marinozzi MC, Berre S, Terulla C, et al. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J Med Virol 2008;80: 716–23.
- Esper FP, Spahlinger T, Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J Infect 2011;63:260–6.
- Goka EA, Vallely PJ, Mutton KJ, Klapper PE. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect 2015;143: 37–47.
- Goka E, Vallely P, Mutton K, Klapper P. Influenza A viruses dual and multiple infections with other respiratory viruses and risk of hospitalisation and mortality. Influenza Other Respir Viruses 2013;7:1079–87.
- Mazur NI, Bont L, Cohen AL, Cohen C, von Gottberg A, Groome MJ, et al. Severity of respiratory syncytial virus lower respiratory tract infection with viral coinfection in HIV-uninfected children. Clin Infect Dis 2017;64: 443–50.
- Hanson KE, Azar MM, Banerjee R, Chou A, Colgrove RC, Ginocchio CC, et al. Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA's diagnostics committee. Clin Infect Dis 2020;71:2744–51.
- Brummer LE, Katzenschlager S, Gaeddert M, Erdmann C, Schmitz S, Bota M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Med 2021;18:e1003735.
- Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 2021;3: CD013705.
- Pray IW, Ford L, Cole D, Lee C, Bigouette JP, Abedi GR, et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September-October 2020. MMWR Morb Mortal Wkly Rep 2021;69:1642–7.
- Hayden MK, Hanson KE, Englund JA, Lee F, Lee MJ, Loeb M, et al. IDSA guidelines on the diagnosis of COVID-19: Antigen testing; Version 2.0.0. Infectious Diseases Society of America; 2022. https://www.idsociety.org/ practice-guideline/covid-19-guideline-antigen testing/ (Accessed February 2023).
- CDC. Guidance for antigen testing for SARS-CoV-2 for healthcare providers testing individuals in the community. Updated April 4, 2022. https://stacks.cdc.gov/view/cdc/ 115045 (Accessed February 2024).
- Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis 2021;73:e2861–6.
- 90. McKay SL, Tobolowsky FA, Moritz ED, Hatfield KM, Bhatnagar A, LaVoie SP, et al. Performance evaluation of serial SARS-CoV-2 rapid antigen testing during a nursing home outbreak. Ann Intern Med 2021;174: 945–51.
- Prince-Guerra JL, Almendares O, Nolen LD, Gunn JKL, Dale AP, Buono SA, et al. Evaluation of Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb Mortal Wkly Rep 2021;70:100–5.
- Hardick J, Gallagher N, Sachithanandham J, Fall A, Siddiqui Z, Pekosz A, et al. Evaluation of four point of care (POC) antigen assays for the detection of the SARS-CoV-2 variant omicron. Microbiol Spectr 2022; 10:e0102522.
- Uyeki TM, Bernstein HH, Bradley JS, Englund JA, File TM Jr, Fry AM, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa. Clin Infect Dis 2018;68:e147.
- CDC. Algorithm to assist in the interpretation of influenza testing results and clinical decision-making during periods when influenza viruses are circulating in the community. https://www.cdc.gov/flu/professionals/ diagnosis/algorithm-results-circulating.htm (Accessed February 2023).
- Chartrand C, Tremblay N, Renaud C, Papenburg J. Diagnostic accuracy of rapid antigen detection tests for respiratory syncytial virus infection: systematic review and meta-analysis. J Clin Microbiol 2015;53:3738–49.
- Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med 2020;382:2372–4.
- Dioverti MV, Gaston DC, Morris CP, Huff CA, Jain T, Jones R, et al. Combination therapy with Casirivimab/ Imdevimab and Remdesivir for protracted SARS-CoV-2 infection in B-cell-depleted patients. Open Forum Infect Dis 2022;9:ofac064.
- Mowrer CT, Creager H, Cawcutt K, Birge J, Lyden E, Van Schooneveld TC, et al. Evaluation of cycle threshold values at deisolation. Infect Control Hosp Epidemiol 2022;43:794–6.
- Rao SN, Manissero D, Steele VR, Pareja J. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther 2020;9:573–86.
- Shenoy S. SARS-CoV-2 (COVID-19), viral load and clinical outcomes; lessons learned one year into the pandemic: a systematic review. World J Crit Care Med 2021;10: 132–50.
- Erard V, Huang ML, Ferrenberg J, Nguy L, Stevens-Ayers TL, Hackman RC, et al. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin Infect Dis 2007;45: 958–65.
- Lalueza A, Folgueira D, Muñoz-Gallego I, Trujillo H, Laureiro J, Hernández-Jiménez P, et al. Influence of viral load in the outcome of hospitalized patients with influenza virus infection. Eur J Clin Microbiol Infect Dis 2019;38:667–73.
- Song C, Li Y, Zhou Y, Liang L, Turtle L, Wang F, et al. Enterovirus genomic load and disease severity among children hospitalised with hand, foot and mouth disease. EBioMedicine 2020;62:103078.
- McKay B, Ebell M, Billings WZ, Dale AP, Shen Y, Handel A. Associations between relative viral load at diagnosis and influenza A symptoms and recovery. Open Forum Infect Dis 2020;7:ofaa494.
- Saleeby CM E, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 2011;204: 996–1002.
- Sola I, Almazan F, Zuniga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2015;2:265–88.
- Dagotto G, Mercado NB, Martinez DR, Hou YJ, Nkolola JP, Carnahan RH, et al. Comparison of subgenomic and total RNA in SARS-CoV-2 challenged rhesus macaques. J Virol 2021;95:e02370-20.
- Perera R, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 2020;26:2701–4.
- Dimcheff DE, Valesano AL, Rumfelt KE, Fitzsimmons WJ, Blair C, Mirabelli C, et al. Severe acute respiratory syndrome coronavirus 2 total and subgenomic RNA viral load in hospitalized patients. J Infect Dis 2021;224: 1287–1293.
- Deming ME, Dong TQ, Agrawal V, Mills MG, Huang MLW, Greninger AL, et al. Detection and kinetics of subgenomic severe acute respiratory syndrome coronavirus 2 RNA viral load in longitudinal diagnostic RNA-positive samples. J Infect Dis 2022;226:788–96.
- Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis 2011;52(Suppl 4):S284–9.
- Uyeki TM, Bernstein HH, Bradley JS, Englund JA, File TM, Fry AM, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal Influenzaa. Clin Infect Dis 2019;68:895–902.
- Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 9th Ed. Philadelphia (PA): Elsevier; 2020.
- Lee JJ, Verbakel JY, Goyder CR, Ananthakumar T, Tan PS, Turner PJ, et al. The clinical utility of point-of-care tests for influenza in ambulatory care: a systematic review and meta-analysis. Clin Infect Dis 2019;69:24–33.
- Clark TW, Beard KR, Brendish NJ, Malachira AK, Mills S, Chan C, et al. Clinical impact of a routine, molecular, point-of-care, test-and-treat strategy for influenza in adults admitted to hospital (FluPOC): a multicentre, open-label, randomised controlled trial. Lancet Respir Med 2021;9:419–29.
- Green DA, Hitoaliaj L, Kotansky B, Campbell SM, Peaper DR. Clinical utility of on-demand multiplex respiratory pathogen testing among adult outpatients. J Clin Microbiol 2016;54:2950–5.
- Claus JA, Hodowanec AC, Singh K. Poor positive predictive value of influenza-like illness criteria in adult transplant patients: a case for multiplex respiratory virus PCR testing. Clin Transplant 2015;29:938–43.
- van Someren Greve F, Ong DS, Cremer OL, Bonten MJ, Bos LD, de Jong MD, et al. Clinical practice of respiratory virus diagnostics in critically ill patients with a suspected pneumonia: a prospective observational study. J Clin Virol 2016;83:37–42.
- Cousin M, Molinari N, Foulongne V, Caimmi D, Vachier I, Abely M, Chiron R. Rhinovirus-associated pulmonary exacerbations show a lack of FEV1 improvement in children with cystic fibrosis. Influenza Other Respir Viruses 2016;10:109–12.
- Ginocchio CC. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens. Clin Infect Dis 2011;52(Suppl 4):S312–25.
- Weiss ZF, Cunha CB, Chambers AB, Carr AV, Rochat C, Raglow-Defranco M, et al. Opportunities revealed for antimicrobial stewardship and clinical practice with implementation of a rapid respiratory multiplex assay. J Clin Microbiol 2019;57:e00861-19.
- Rogers BB, Shankar P, Jerris RC, Kotzbauer D, Anderson EJ, Watson JR, et al. Impact of a rapid respiratory panel test on patient outcomes. Arch Pathol Lab Med 2015; 139:636–41.
- Rappo U, Schuetz AN, Jenkins SG, Calfee DP, Walsh TJ, Wells MT, et al. Impact of early detection of respiratory viruses by multiplex PCR assay on clinical outcomes in adult patients. J Clin Microbiol 2016;54:2096–103.
- Brendish NJ, Malachira AK, Armstrong L, Houghton R, Aitken S, Nyimbili E, et al. Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir Med 2017;5:401–11.
- Rao S, Lamb MM, Moss A, Mistry RD, Grice K, Ahmed W, et al. Effect of rapid respiratory virus testing on antibiotic prescribing among children presenting to the emergency department with acute respiratory illness: a randomized clinical trial. JAMA Netw Open 2021;4:e2111836.
- May L, Tatro G, Poltavskiy E, Mooso B, Hon S, Bang H, Polage C. Rapid multiplex testing for upper respiratory pathogens in the emergency department: a randomized controlled trial. Open Forum Infect Dis 2019;6:ofz481.
- Mattila S, Paalanne N, Honkila M, Pokka T, Tapiainen T. Effect of point-of-care testing for respiratory pathogens on antibiotic use in children: a randomized clinical trial. JAMA Netw Open 2022;5:e2216162.
- Aitken C, Jeffries DJ. Nosocomial spread of viral disease. Clin Microbiol Rev 2001;14:528–46.
- Schreckenberger PC, McAdam AJ. Point-counterpoint: large multiplex PCR panels should be first-line tests for detection of respiratory and intestinal pathogens. J Clin Microbiol 2015;53:3110–5.
- Quer J, Colomer-Castell S, Campos C, Andres C, Pinana M, Cortese MF, et al. Next-generation sequencing for confronting virus pandemics. Viruses 2022;14:600.
- Greninger AL, Zerr DM, Qin X, Adler AL, Sampoleo R, Kuypers JM, et al. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J Clin Microbiol 2017;55:177–82.
- Sammons JS, Graf EH, Townsend S, Hoegg CL, Smathers SA, Coffin SE, et al. Outbreak of adenovirus in a neonatal intensive care unit: critical importance of equipment cleaning during inpatient ophthalmologic examinations. Ophthalmology 2019;126:137–43.
- Graitcer SB, Gubareva L, Kamimoto L, Doshi S, Vandermeer M, Louie J, et al. Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg Infect Dis 2011;17: 255–7.
- Thorburn F, Bennett S, Modha S, Murdoch D, Gunson R, Murcia PR. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol 2015;69:96–100.
- Vengesai A, Midzi H, Kasambala M, Mutandadzi H, Mduluza-Jokonya TL, Rusakaniko S, et al. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst Rev 2021;10:155.
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 2020;6:CD013652.
- 1Collaborators GL. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 2017;17:1133–61.
- Hay AD, Heron J, Ness A, team As. The prevalence of symptoms and consultations in pre-school children in the avon longitudinal study of parents and children (ALSPAC): a prospective cohort study. Fam Pract 2005; 22:367–74.
- Ramaekers K, Keyaerts E, Rector A, Borremans A, Beuselinck K, Lagrou K, Van Ranst M. Prevalence and seasonality of six respiratory viruses during five consecutive epidemic seasons in Belgium. J Clin Virol 2017;94:72–8.
- Schroeder AR, Ralston SL. Viral testing for pediatric respiratory infections: why precise diagnoses do not always translate to patient benefit. JAMA 2017;318:472–3.
- Kitano T, Nishikawa H, Suzuki R, Onaka M, Nishiyama A, Kitagawa D, et al. The impact analysis of a multiplex PCR respiratory panel for hospitalized pediatric respiratory infections in Japan. J Infect Chemother 2020; 26:82–5.
- Lee BR, Hassan F, Jackson MA, Selvarangan R. Impact of multiplex molecular assay turnaround-time on antibiotic utilization and clinical management of hospitalized children with acute respiratory tract infections. J Clin Virol 2019;110:11–6.
- Subramony A, Zachariah P, Krones A, Whittier S, Saiman L. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J Pediatr 2016;173: 196–201e2.
- Nikolopoulou GB, Maltezou HC. COVID-19 in children: where do we stand? Arch Med Res 2022;53:1–8.
- 145. Kociolek LK, Muller WJ, Yee R, Dien Bard J, Brown CA, Revell PA, et al. Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS-CoV-2 infection in pediatric hospital testing programs. J Clin Microbiol 2020;59:e02593-20.
- Laporte L, Hermetet C, Jouan Y, Gaborit C, Rouve E, Shea KM, et al. Ten-year trends in intensive care admissions for respiratory infections in the elderly. Ann Intensive Care 2018;8:84.
- Liao RS, Appelgate DM, Pelz RK. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility for the elderly in Oregon. J Clin Virol 2012;53:171–3.
- Czaja CA, Miller L, Alden N, Wald HL, Cummings CN, Rolfes MA, et al. Age-related differences in hospitalization rates, clinical presentation, and outcomes among older adults hospitalized with influenza-U.S. influenza hospitalization surveillance network (FluSurv-NET). Open Forum Infect Dis 2019; 6:ofz225.
- Centers for Disease C, Prevention. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb Mortal Wkly Rep 2010;59: 1057–62.
- Reed C, Chaves SS, Daily Kirley P, Emerson R, Aragon D, Hancock EB, et al. Estimating influenza disease burden from population-based surveillance data in the United States. PLoS One 2015;10:e0118369.
- Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373:415–27.
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 2005;352:1749–59.
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 2018;391:1285–300.
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017;390:946–58.
- Chemaly RF, Shah DP, Boeckh MJ. Management of respiratory viral infections in hematopoietic cell transplant recipients and patients with hematologic malignancies. Clin Infect Dis 2014;59(Suppl 5): S344–51.
- Pilie P, Werbel WA, Jt R, Shu X, Schaubel D, Gregg KS. Adult patients with respiratory syncytial virus infection: impact of solid organ and hematopoietic stem cell transplantation on outcomes. Transpl Infect Dis 2015; 17:551–7.
- Shah DP, Ghantoji SS, Shah JN, El Taoum KK, Jiang Y, Popat U, et al. Impact of aerosolized ribavirin on mortality in 280 allogeneic haematopoietic stem cell transplant recipients with respiratory syncytial virus infections. J Antimicrob Chemother 2013;68:1872–80.
- Hijano DR, Maron G, Hayden RT. Respiratory viral infections in patients with cancer or undergoing hematopoietic cell transplant. Front Microbiol 2018; 9:3097.
- Kim YJ, Boeckh M, Englund JA. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin Respir Crit Care Med 2007;28:222–42.
- Schanzer DL, Saboui M, Lee L, Nwosu A, Bancej C. Burden of influenza, respiratory syncytial virus, and other respiratory viruses and the completeness of respiratory viral identification among respiratory inpatients, Canada, 2003-2014. Influenza Other Respir Viruses 2018;12:113–21.
- Ortiz JR, Neuzil KM, Shay DK, Rue TC, Neradilek MB, Zhou H, et al. The burden of influenza-associated critical illness hospitalizations. Crit Care Med 2014;42:2325–32.
- Chu HY, Englund JA, Huang D, Scott E, Chan JD, Jain R, et al. Impact of rapid influenza PCR testing on hospitalization and antiviral use: a retrospective cohort study. J Med Virol 2015;87:2021–6.
- Yeung V A, Thapa K, Rawlinson W, Georgiou A, Post JJ, Overton K. Differences in antibiotic and antiviral use in people with confirmed influenza: a retrospective comparison of rapid influenza PCR and multiplex respiratory virus PCR tests. BMC Infect Dis 2021;21:321.
- Paz V, D'Agostino ML, Garibaldi F, Orellana R, Paniagua M, Santillan A. Multiplex PCR in the empirical antibiotic treatment of patients with SARS-CoV-2 and bacterial respiratory superinfection. Infect Prev Pract 2022; 4:100227.
- CDC. Influenza (Flu). https://www.cdc.gov/flu/ professionals/diagnosis/testing-guidance-for-clinicians. htm (Accessed September 2023).
- CDC. Respiratory syncytial virus (RSV). https://www.cdc. gov/rsv/clinical/index.html (Accessed September 2023).
- Messacar K, Parker SK, Todd JK, Dominguez SR. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol 2017;55: 715–23.
- Kressel A, Cheatham M, Chang A. Diagnostic stewardship of respiratory pathogen panel utilization. Antimicrob Steward Healthc Epidemiol 2021;1:1.
- Poelman R, der Meer JV, der Spek CV, Riezebos-Brilman A, Knoester M, Leer-Buter CV, et al. Improved diagnostic policy for respiratory tract infections essential for patient management in the emergency department. Future Microbiol 2020;15:623–32.
- Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007;44(Suppl 2): S27–72.
- Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, Li Y, et al. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin Microbiol Rev 2019;32:e00042-18.
- American Society for Microbiology. Clinical utility of multiplex tests for respiratory and GI pathogens. https:// asm.org/guideline/clinical-utility-of-multiplex-tests-forrespirator (Accessed September 2023).
- Hanson KE, Caliendo AM, Arias CA, Hayden MK, Englund JA, Lee MJ, et al. The Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: molecular diagnostic testing. Clin Infect Dis 2021:ciab048.
- Committee On Infectious D. Recommendations for prevention and control of influenza in children, 2021– 2022. Pediatrics 2021;148:e2021053745.
- American Academy of Pediatrics. COVID-19 testing guidance. https://www.aap.org/en/pages/2019-novelcoronavirus- covid-19-infections/clinical-guidance/covid- 19-testing-guidance/ (Accessed September 2023).
- Evans SE, Jennerich AL, Azar MM, Cao B, Crothers K, Dickson RP, et al. Nucleic acid-based testing for non-influenza viral pathogens in adults with suspected community-acquired pneumonia. An Official American Thoracic Society Clinical Practice Guideline. Am J Respir Crit Care Med 2021;203:1070–87.
- von Lilienfeld-Toal M, Berger A, Christopeit M, Hentrich M, Heussel CP, Kalkreuth J, et al. Community acquired respiratory virus infections in cancer patients-guideline on diagnosis and management by the Infectious Diseases Working Party of the German society for haematology and medical oncology. Eur J Cancer 2016; 67:200–12.
- Hanson KE, Caliendo AM, Arias CA, Englund JA, Hayden MK, Lee MJ, et al. Infectious diseases society of America Guidelines on the Diagnosis of COVID-19:serologic testing. Clin Infect Dis 2020:ciaa1343.
- Uhteg K, Jarrett J, Richards M, Howard C, Morehead E, Geahr M, et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol 2020;127:104384.
- Aden TA, Blevins P, York SW, Rager S, Balachandran D, Hutson CL, et al. Rapid diagnostic testing for response to the monkeypox outbreak—laboratory response network, United States, May 17-June 30, 2022. MMWR Morb Mortal Wkly Rep 2022;71:904–7.
- Greninger AL. Test it earlier, result it faster, makes us stronger: how rapid viral diagnostics enable therapeutic success. Curr Opin Virol 2021;49:111–6.

